Recent research has unveiled a striking complexity within single-celled organisms—specifically bacteria and archaea—that challenges previous assumptions regarding DNA packaging proteins. Historically, histones were thought to be exclusive to more complex life forms. However, emerging findings now reveal that these proteins are not only present within archaeal and bacterial cells but that their forms and functionalities are much more varied than earlier predictions suggested. Such groundbreaking work was spearheaded by Samuel Schwab, a Ph.D. candidate at Leiden University, whose exhaustive exploration into the structures of histones offers a fresh perspective on genetic material organization in prokaryotes.
To understand the significance of this discovery, it is essential to grasp the role of histones within cellular biology. DNA, by its very nature, is an overwhelmingly large molecule that cannot simply exist unraveled within the confines of a cell. The role of histones is akin to that of spools for thread—they compact and organize DNA into tightly packed structures. In eukaryotic organisms, histones form nucleosomes, the building blocks of chromatin that enable efficient genetic material management. Schwab’s work extends this understanding to single-celled organisms, revealing that their histones also function in similar yet diverse capacities.
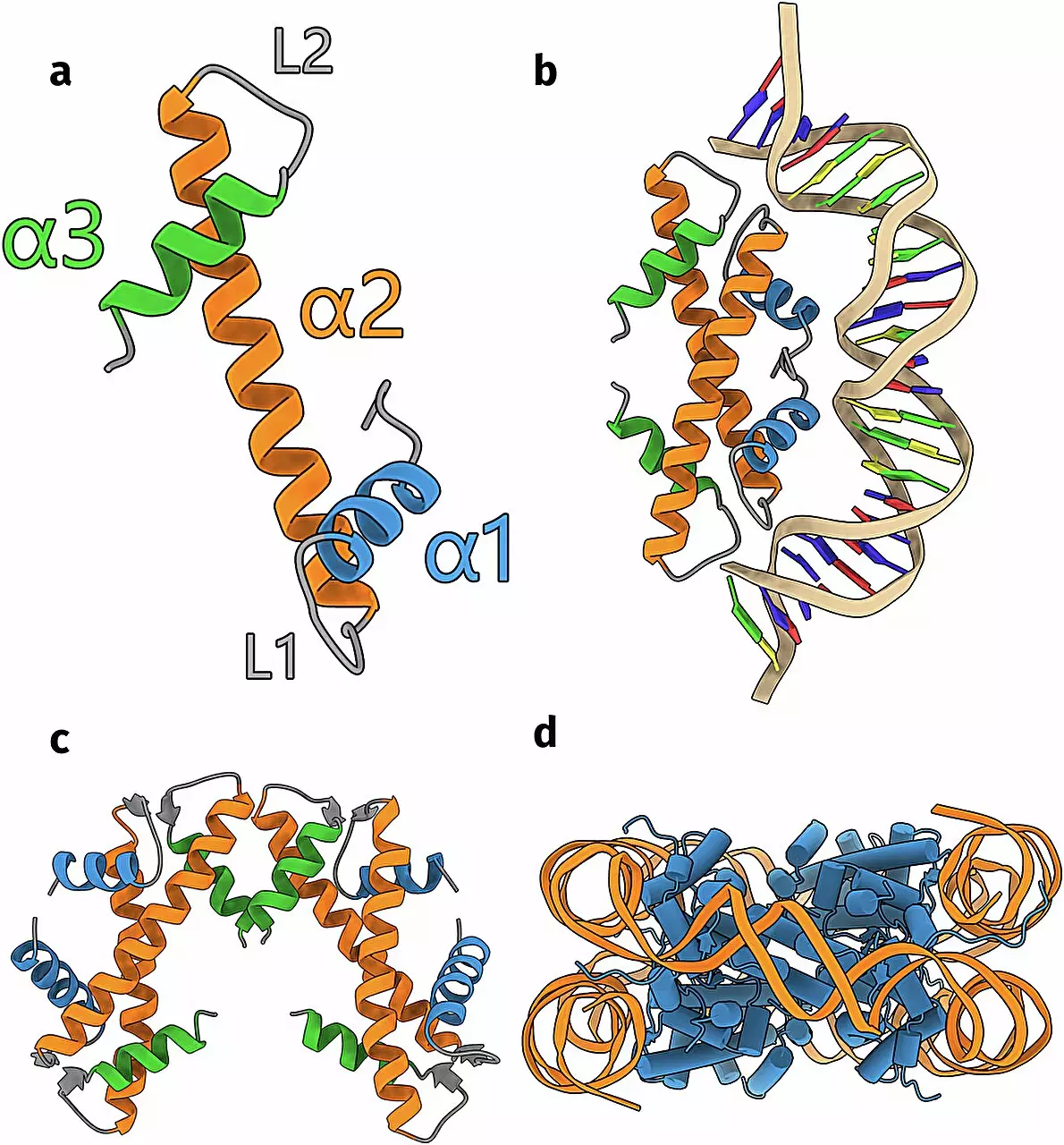
The research took a monumental turn with the introduction of AlphaFold, a state-of-the-art artificial intelligence algorithm known for its ability to predict protein structures from genetic sequences. Schwab and his team harnessed this technology to analyze an extensive protein database, identifying approximately 6,000 sequences potentially coding for unknown histones within archaea and bacteria. This intersection of biology and technology highlights the transformative potential of AI in scientific research, demonstrating how advanced computational tools can elucidate complex biological phenomena that might otherwise remain obscured.
The findings presented by Schwab are both extensive and intricate, identifying seventeen distinct groups of histones. While a portion of these groups aligns with previously cataloged structures, Schwab’s work also highlights the existence of entirely novel histone forms. This staggering diversity invites deeper inquiries into the specific roles these proteins play within their respective organisms and the mechanisms through which they interact with DNA. Crucially, the research not only confirms the predictive accuracy of AlphaFold but also affirms the validity of the theoretical framework upon which these protein structures were interpreted.
Perhaps one of the most thought-provoking aspects of Schwab’s research is the suggestion that some histones do not merely engage with DNA but also have evolutionary adaptations that allow them to bind with cellular membranes. This revelation not only broadens the scope of histone functionality but also raises questions about the evolutionary pressures that shaped these interactions. What roles do these membrane-binding histones serve within the cellular framework? This critical inquiry opens new avenues for investigation into the molecular biology of prokaryotes, particularly concerning the interplay of genetic organization and cellular architecture.
The work of Schwab and the Dame lab contributes significantly to our understanding of DNA organization, particularly in light of evolutionary biology. Insights into how different organisms manage their genetic materials may provide critical interpretations of DNA data and measurements, guiding future research endeavors. Remus Dame, Schwab’s supervisor, further emphasizes the necessity of continued inquiry into this area, as a more robust understanding of histone diversity will enhance our comprehension when interpreting cellular phenomena.
As Schwab aptly notes, a substantial amount remains to be learned regarding the roles of these histones within their cellular environments. The research serves as a foundational piece, revealing the intricate relationships that exist between single-celled organisms and their genetic machinery. With continued advances in analytical methodologies and heightened interest in the less explored areas of microbiology, the next phase of research holds promising potential. As the scientific community embraces this newfound complexity, our understanding of the microscopic world will undoubtedly become richer and more nuanced.